Scientists at MIT McGovern Brain Institute and an extensive institute at MIT and Harvard University have redesigned a compact RNA-guided enzyme that the enzymes found in bacteria are effective, programmable editors for human DNA.
The proteins they create, called Novaiscb, can be adapted to make precise changes to the genetic code, regulate the activity of a specific gene, or perform other editing tasks. Because its small size can simplify delivery to cells, the developers of Novaiscb say it is a promising candidate for developing gene therapies for treating or preventing diseases.
The study was led by James and Patricia Poitras Professor of Neuroscience at MIT, Feng Zhang, who is also a researcher at the McGovern Institute and Howard Hughes Medical Institute and a core member of the Broad Institute. Zhang and his team report this month’s open work in their diary Natural Biotechnology.
Novaiscb originates from a bacterial DNA cutter belonging to a protein called ISCB, Zhang’s lab discovered in 2021. ISCBS is a type of Omega system and is an evolutionary ancestor of Cas9, part of the bacterial CRISPR system that Zhang and other bacterial CRISPR systems that have developed into powerful genome editing tools. Like CAS9, ISCB enzymes cleave DNA at sites specified in the RNA guideline. By reprogramming the guide, researchers can redirect the enzyme to its targeting sequence of choice.
The ISCB caught the team’s attention not only because they have the key features of CRISPR’s DNA cutting into CAS9, but also because they are one-third of their size. This is an advantage for potential gene therapies: compact tools are easier to pass to cells, while using small enzymes, researchers will have greater flexibility to patch, potentially adding new features without creating too large clinically used tools.
From the initial study of ISCB, researchers in Zhang’s lab learned that some members of the family could cut off DNA targets in human cells. However, none of the bacterial protein works well enough to be deployed through treatment: the team must modify the ISCB to ensure it can effectively edit targets in human cells without disturbing the rest of the genome.
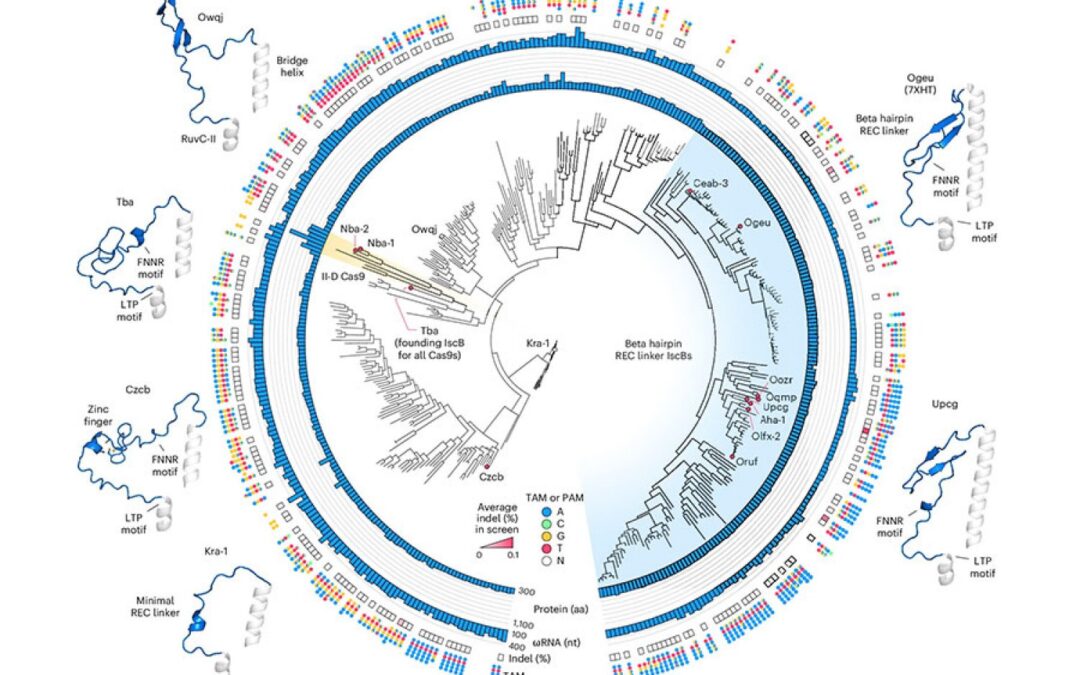
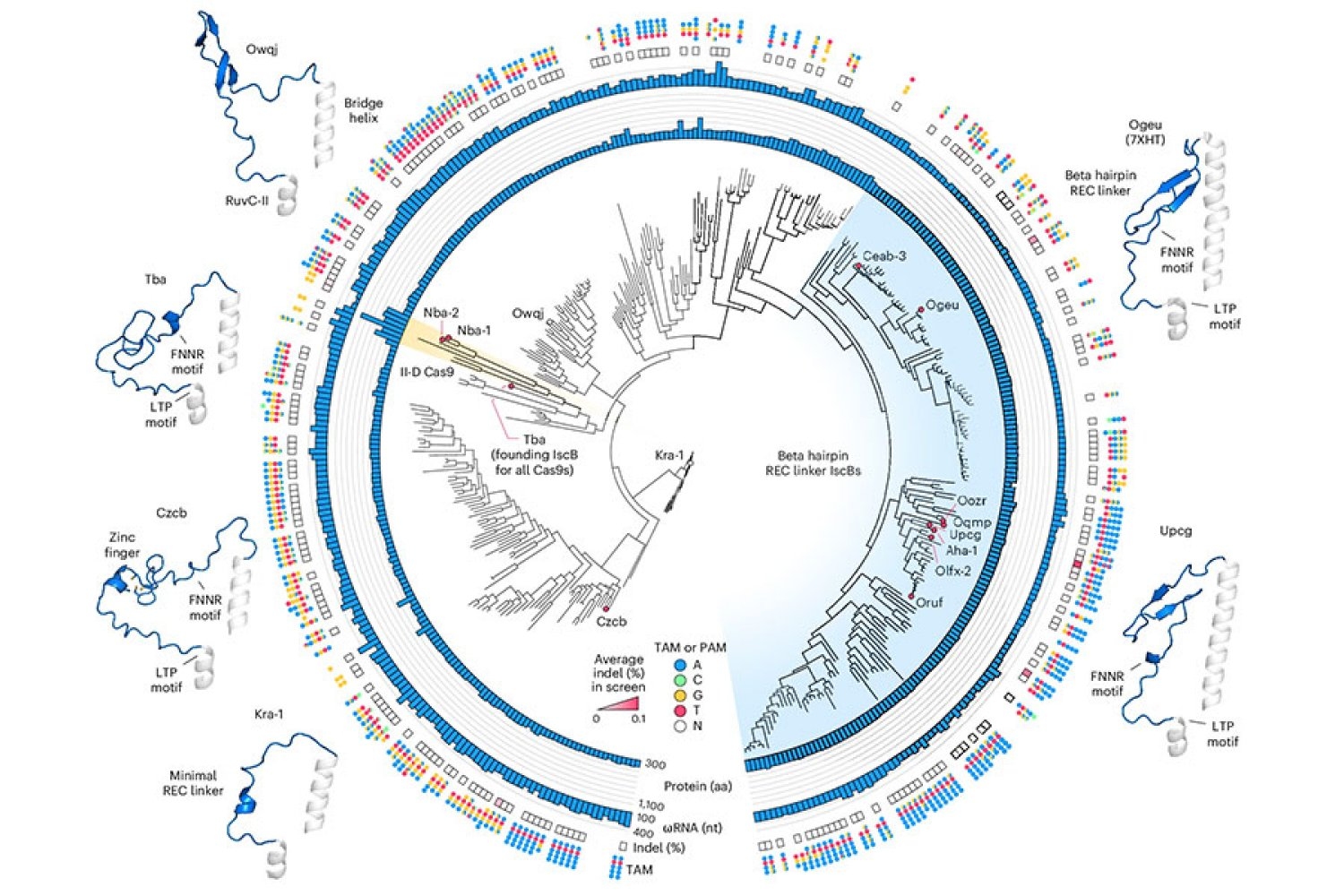
To start this engineering process, Zhang’s graduate student Soumya Kannan is now a junior fellow at the Harvard Fellows Association, and Postdoc Shiyou Zhu first searched for an ISCB that could be a good starting point. They tested nearly 400 different ISCB enzymes that can be found in bacteria. Ten are able to edit DNA in human cells.
Even the most active people need to enhance it to make it a useful genome editing tool. The challenge would be to increase the activity of the enzyme, but only on the sequences specified in its RNA guidelines. If the enzyme becomes more active, but not bad, it will cut DNA in unexpected places. “The key is to balance the activity and specificity improvements at the same time,” Zhu explained.
Zhu noted that bacterial ISCB targets its target sequences through relatively short RNA guidelines, which makes it difficult to limit the activity of the enzyme to specific parts of the genome. If the ISCB can be designed to accommodate longer guidelines, there is less chance of action on sequences that exceed their intended goals.
To optimize ISCB for human genome editing, the team used information from graduate student Han Altae-Tran, now a postdoctoral fellow at the University of Washington, to understand the diversity of bacterial ISCBs and how they develop. For example, researchers point out that the ISCBs that function in human cells include a segment they call rec, which does not exist in other ISCBs. They suspect that the enzyme may require the fragment to interact with DNA in human cells. When they looked at the region closely, structural modeling showed that by slightly expanding part of the protein, REC might also enable ISCB to recognize longer RNA guidelines.
Based on these observations, the team tried partial exchange of REC domains from different ISCBs and CAS9 to evaluate how each change affects the function of the protein. Under their guidance on how ISCB and CAS9 interact with DNA and its RNA guidelines, the researchers made additional changes to optimize efficiency and specificity.
Finally, they produced a protein called Novaiscb that was more than 100 times active in human cells, 100 times higher than ISCB that started using, and showed good specificity for its target.
Kannan and Zhu constructed and screened hundreds of new ISCBs before reaching Novaiscb, and every change they made to the original protein was strategic. Their efforts are guided by the team’s understanding of the natural evolution of ISCB and the prediction of how each change will affect protein structure, and are made using an artificial intelligence tool called Alphafold2. This reasonable engineering approach greatly accelerated the team’s ability to identify proteins that are required to function compared to traditional methods that introduce random changes into proteins and screen for their effects.
The team shows that Novaiscb is a good scaffolding for various genome editing tools. “Its biochemical function is very similar to that of CAS9, which makes it easy to transplant tools that have been optimized with Cas9 scaffolding,” Canan said. With different modifications, the researchers used Novaisbb to replace specific letters of DNA code in human cells and alter the activity of targeted genes.
Importantly, news-based tools are compact enough to easily package in a single adeno-associated virus (AAV), the most commonly used vector for safe delivery of gene therapy to patients. Because they are larger, tools developed using CAS9 may require more complex delivery strategies.
Zhang’s team demonstrated the therapeutic use potential of Novaiscb, creating a tool called Omegaoff that adds chemical markers to DNA to dial the activity of a specific gene. They programmed Omegaoff to inhibit genes involved in cholesterol regulation and then used AAV to deliver the system to the liver of mice, resulting in a sustained decrease in cholesterol levels in the animal’s blood.
The team predicts that NovaIscb can be used to localize genome editing tools to most human genes and looks forward to seeing how other labs deploy new technologies. They also hope that others will adopt their evolution-guided approach to rational protein engineering. “Nature has such diversity, and its systems have different advantages and disadvantages,” Zhu said. “By understanding this natural diversity, we can make the system we try to design better and better.”
The study was funded in part by K. Lisa Yang and Hock E. Tan Center for Molecular Therapy, Broad Institute Programmable Therapy Donors, Pershing Square Founders, William Ackman, Neri Oxman, Phillips Family, Phillips Family and J. and Poitras at MIT.

 1005 Alcyon Dr Bellmawr NJ 08031
1005 Alcyon Dr Bellmawr NJ 08031