For patients with inflammatory bowel disease, antibiotics can be a double-edged sword. Broad-spectrum drugs, often prescribed for intestinal outbreaks, can kill useful microorganisms together with harmful microorganisms, and sometimes worsen symptoms over time. When fighting intestinal inflammation, you don’t always want to bring a sledgehammer for a tool.
Researchers at the MIT Computer Science and Artificial Intelligence Laboratory (CSAIL) and McMaster University have identified a new compound that adopts a more targeted approach. The molecule is called intestoloprotein, inhibiting a group of bacteria associated with the Crohn’s disease outbreak while leaving the rest of the microbiome completely intact. The team used the generated AI model to map how the compounds work, a process that usually takes years but accelerates to several months here.
McMaster’s new paper, assistant professor of biochemistry and biomedical sciences, research branch of MIT’s Abdul Latif Jameel Jameel Clinic, is about the core challenges in antibiotic development. “The problem is not to find the molecules that kill bacteria in dishes – we have been able to do that for a long time. One major obstacle is figuring out what these molecules actually do in bacteria. Without a detailed understanding, these early stage antibiotics cannot be developed into safe and effective treatments for patients. ”
Intestinal Lorene protein is a big step towards precise antibiotics: treatments designed to knock out only bacteria that cause trouble. In a mouse model of Crones-like inflammation, the drug zeroes on ON They show chillThis is a gut bacteria that can worsen the flares while keeping most other microbial residents out of touch. Mice given intestoloprotein maintain their microbiome faster than mice treated with vancomycin, a common antibiotic.
Fixing the mechanism of action of a drug, the molecular targets it binds in bacterial cells usually requires years of hard experimentation. Stokes’ lab used a high-throughput screening method to detect intestinal lorraline, but determined its target was a bottleneck. Here, the team turned to the generative AI model Diffdock developed at CSAIL by MIT PhD student Gabriele Corso and MIT professor Regina Barzilay.
Diffdock aims to predict how small molecules fit into protein binding bags, a well-known difficult problem in structural biology. Traditional docking algorithms often produce noisy results by using scoring rules to search for possible directions. Instead, Diffdock uses docking as a framework for probabilistic reasoning problems: the diffusion model iteratively perfects the guess until it converges in the most likely binding mode.
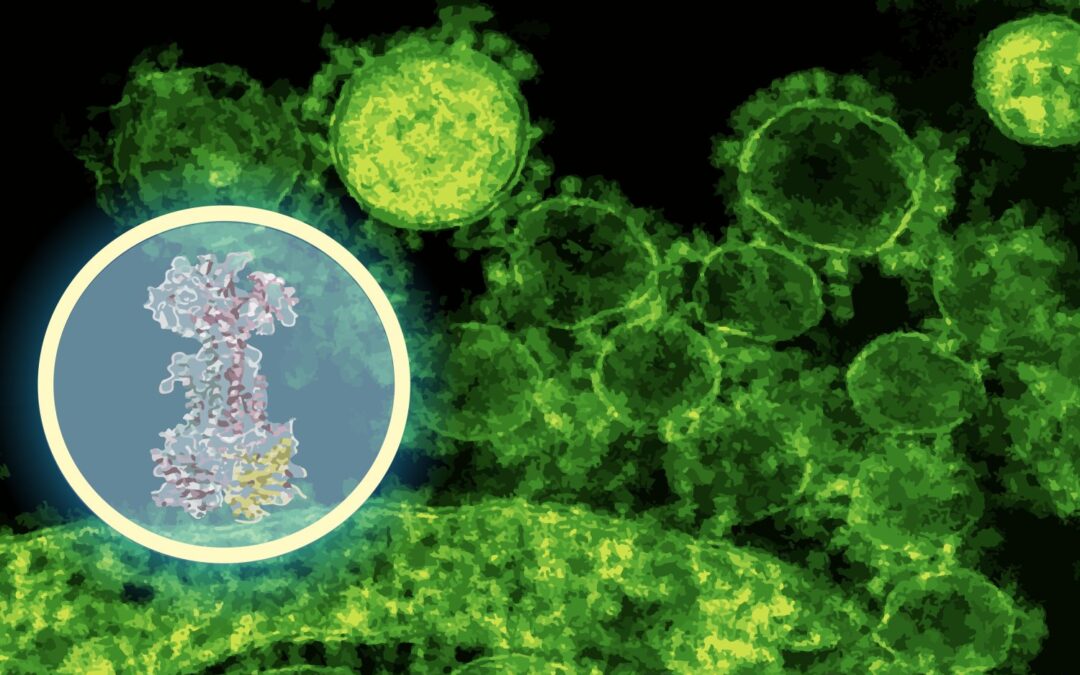
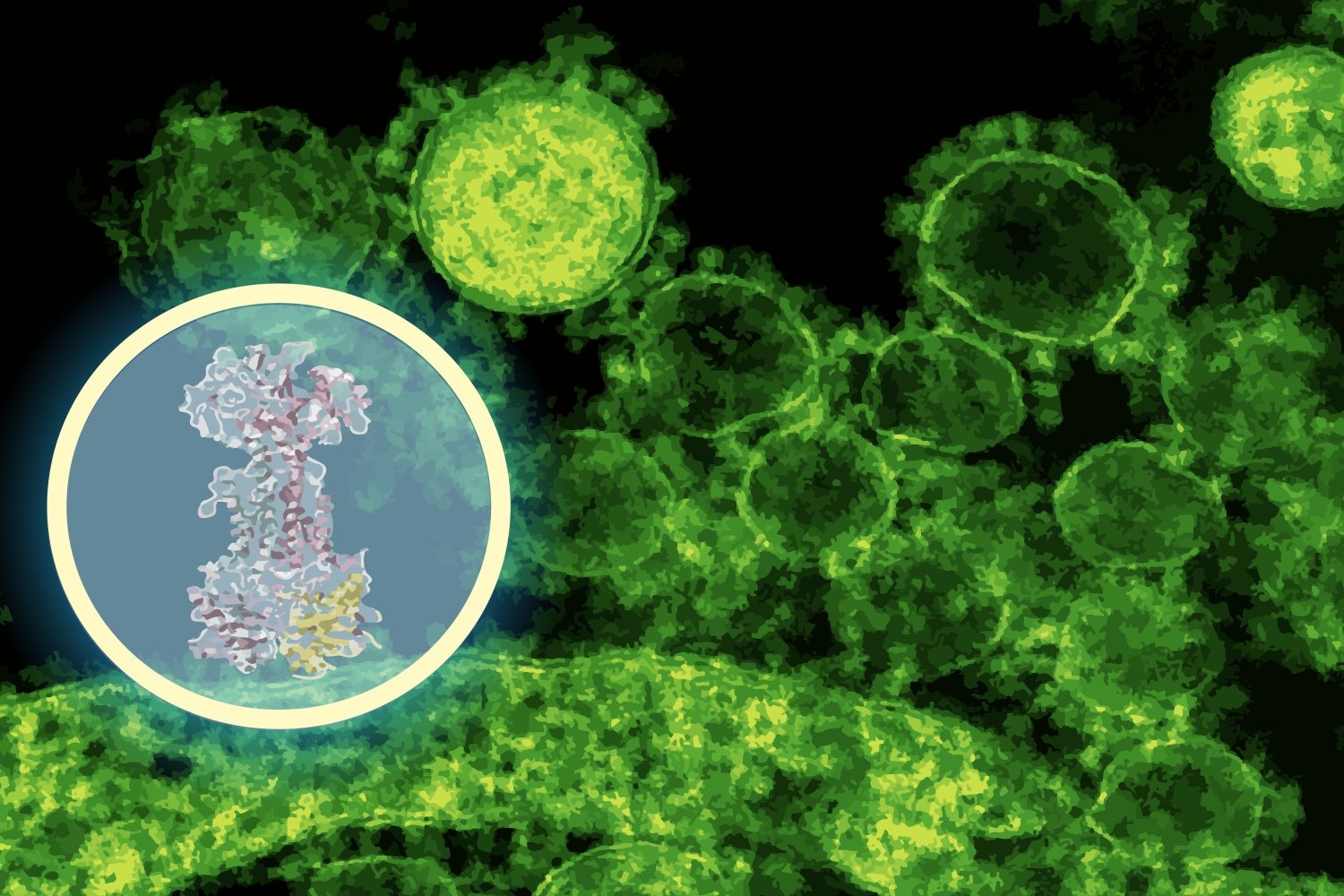
“In just a few minutes, the model predicted that intestinal lorolol protein binds to a protein complex called LOLCDE, which is essential for transporting lipoproteins in certain bacteria,” Barzilay said. “That’s a very specific leader – one can guide the experiment, not replace the experiment.”
Stokes’ team then put the prediction in the test. Using Diffdock predictions as experimental GPs, they evolve first E. coli In the lab, the lab revealed changes in the DNA mapping of the mutant to LOLCDE, which is where Diffdock predicts intestinal lorolorin binding. They also performed RNA sequencing to view bacterial genes that were turned on or off when exposed to the drug and used CRISPR to selectively knock down the expression of the expected target. These laboratory experiments have revealed disruptions in pathways associated with lipoprotein transport, exactly what Diffdock predicts.
“When you see the computational model and wet lab data pointing to the same mechanism, that is when you start to believe that you have figured out something,” Stokes said.
For Barzilay, the project highlights changes in how AI is used in life sciences. “Many of the use of AI in drug discovery is about searching the chemical space to identify new molecules that may be active,” she said. “What we are showing here is that AI can also provide mechanical explanations, which is essential for transferring molecules through the development of pipelines.”
This distinction is important because mechanism of action research is often a major limiting step in drug development. The traditional method can take 18 months to two years or more and costs millions of dollars. In this case, the MIT-MCMASTER team cut the schedule to about six months with a small percentage of the cost.
Enterololin is still in its early stages of development, but translation is already in progress. Stokes’ spinning company Stoked Bio has licensed the compound and is optimizing its potential use properties. Early work is also exploring derivatives of molecules that target other resistant pathogens, such as Klebsiella pneumonia. If all goes well, clinical trials may begin in the next few years.
The researchers also see a broader meaning. Stenosis spectral antibiotics have long been sought as a way to treat infections without damage to the microbiome, but they are difficult to detect and verify. Artificial intelligence tools like Diffdock can make the process more practical, quickly enabling a new generation of targeted antimicrobial agents.
For people with Crohn’s disease and other inflammatory bowel disease, the prospect of drugs that can reduce symptoms without destroying the microbiome may mean meaningful improvements in quality of life. From a larger perspective, precision antibiotics may help deal with the growing threat of antibacterial resistance.
“It’s not just this compound that excites me, but the idea that we can start thinking about elucidating the mechanism of action because we can do things faster with AI, human intuition and laboratory experiments,” Stokes said. “This has the potential to change our drug discovery for many diseases, not just Crohn’s.”
Yves Brun, a professor at the University of Montreal, added: “One of the biggest challenges to our health is the increase in antibacterial bacteria that evade our best antibiotics, nor is it involved in the paper. “AI is becoming an important tool for us to fight these bacteria.” This study uses a powerful and elegant combination of AI approaches to determine the mechanism of action of new antibiotic candidates, an important step in its potential development as a therapeutic one. ”
Corso, Barzilay and Stokes wrote the papers with McMaster researchers Denise B. Catacutan, Vian Tran, Jeremie Alexander, Yeganeh Yousefi, Megan Tu, Stewart McLellan and Dominique Tertigas, as well as professors Jakob Magolan, Michael Surette, Eric Brown, Eric Brown and Brian Coombess. Their research was partially supported by the Weston Family Foundation. David Braley Center for Antibiotic Discovery; Canadian Institute of Health Research; Canadian Council on Natural Science and Engineering; M. and M. Heersink; Canadian Institute of Health Research; Ontario Graduate Scholarships; Jameel Clinic; and U.S. Defense Threat Agency discovers medical responses to new and emerging threat programs.
The researchers released sequencing data in public repositories and published Diffdock-L code publicly on GitHub.

 1005 Alcyon Dr Bellmawr NJ 08031
1005 Alcyon Dr Bellmawr NJ 08031